Unit | Tuberculosis Ecology and Evolution

The Tuberculosis Ecology and Evolution Unit studies the causes and consequences of genetic diversity in the Mycobacterium tuberculosis complex (MTBC), the bacteria that cause tuberculosis (TB). We combine various disciplines to study the global diversity of the MTBC, the evolutionary forces that drive this diversity, and the phenotypic consequences of this diversity for the biology and the epidemiology of TB.
Our research comprises three complementary arms:
- The global population structure of the MTBC
- Ecology and evolution of drug-resistant MTBC
- Genomic epidemiology of TB
Partnerships
An important part of this work relies on our long-term partnerships with collaborators in TB-endemic countries. These include the Ifakara Health Institute in Tanzania, the Noguchi Memorial Institute for Medical Research in Ghana, and the National Centre for Tuberculosis and Lung Disease in Georgia.

Sébastien Gagneux
Professor, PhD
Head of Unit
+41612848369
sebastien.gagneux@swisstph.ch
Main Research Areas of the Tuberculosis Ecology and Evolution Unit
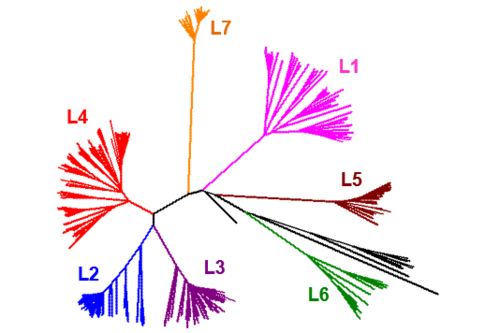
The Global Population Structure of the MTBC
The human-adapted MTBC comprises seven phylogenetic lineages that are associated with different regions of the world. We use comparative whole genome sequencing to study the differences between these lineages and the evolutionary forces shaping this diversity. We combine various –omics technologies with functional assays and epidemiological data to investigate the phenotypic consequences of this diversity. Read more
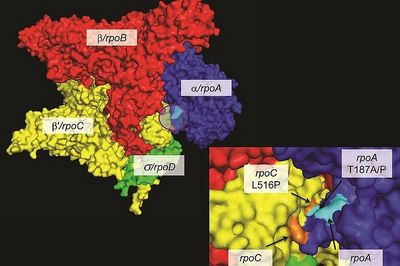
Ecology and Evolution of Drug-Resistant MTBC
Drug resistance poses a growing threat to global health. When drug-resistant bacteria first emerge, they are often less transmissible than susceptible strains – this is because drug resistance in bacteria is often associated with a reduction in Darwinian fitness. However, evolution is a continuous process, and drug-resistant bacteria readily adapt and regain the ability to transmit. This process is mediated by compensatory mutations. Further information
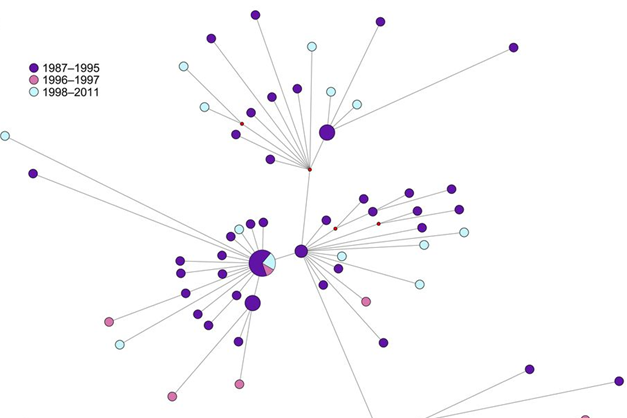
Genomic epidemiology of TB
Recent advances in whole-genome sequencing have revolutionized molecular epidemiological investigation of TB. We use such genomic epidemiological approaches to study the transmission dynamics of TB in Switzerland and in TB-endemic countries. We also explore the micro-evolution of MTBC in individual patients during treatment. More information
Selected Projects
All ProjectsLatest Publications
All PublicationsArbués A, Schmidiger S, Reinhard M, Borrell S, Gagneux S, Portevin D. Soluble immune mediators orchestrate protective in vitro granulomatous responses across Mycobacterium tuberculosis complex lineages. Elife. 2025;13. DOI: 10.7554/eLife.99062
Goig G.A et al. Transmission as a key driver of resistance to the new tuberculosis drugs. N Engl J Med. 2025;392(1):97-99. DOI: 10.1056/NEJMc2404644
Goig G.A et al. Ecology, global diversity and evolutionary mechanisms in the Mycobacterium tuberculosis complex. Nat Rev Microbiol. 2025(in press). DOI: 10.1038/s41579-025-01159-w
March V.F.A et al. Drug-induced differential culturability in diverse strains of Mycobacterium tuberculosis. Sci Rep. 2025;15:3588. DOI: 10.1038/s41598-024-85092-7
Pfurtscheller T et al. Programmatic diagnostic accuracy and clinical utility of Xpert MTB/XDR in patients with rifampicin-resistant tuberculosis in Georgia. Open Forum Infect Dis. 2025;12(2):ofaf022. DOI: 10.1093/ofid/ofaf022
 Amir Banaei Esfahani
Amir Banaei Esfahani
 Sonia Borrell
Sonia Borrell
 Selim Bouaouina
Selim Bouaouina
 Daniela Brites
Daniela Brites
 Galo Goig
Galo Goig
 Sevda Kalkan
Sevda Kalkan
 Chloé Marie Loiseau
Chloé Marie Loiseau
 Valerie March
Valerie March