Group | Diagnostic Development
Diagnostic tests are essential to guide therapy of infectious diseases and to direct control and elimination programs. Although substantial progress has been made for the treatment of many tropical infectious diseases, the development of inexpensive point-of-care diagnostics is lagging behind. For many infections, the available diagnostics are not sufficient to inform physicians on how to adequately manage the patient or address the evolving needs of disease control and elimination.
The Diagnostic Development Group develops and validates novel diagnostic tools from inexpensive point-of-care Rapid diagnostic tests (RDTs) to multi-disease diagnostic surveillance platforms with a focus on characterization of "undifferentiated febrile disease" pathogens. Ongoing projects include the Novel integrated infectious diseases diagnosis and surveillance system (NIIDS) project, where we have developed a urine-based biochemical triage test for assessment of disease severity and are developing a serological microarray test for multiple febrile diseases (Figure 1 below).
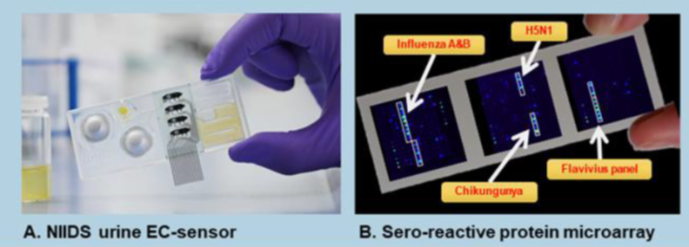
In the Urine Analysis Digital Dipstick (U-ADD) project, we are continuing the work on our urine sensor development. We are incorporating an additional two analytes (creatinine, and albumin) in the sensor and developing it further for the diagnosis of kidney diseases, which are a largely under appreciated common non-communicable diseases in low- and middle-income countries.

We work with partner groups both within Swiss TPH and at external institutes to develop RDTs for diseases of poverty. We have developed a saliva-based test for SARS-CoV-2 detection (completed in 2022) and have two further tests in the pipeline for development; an innovative non-invasive saliva-based influenza RDT and a fingerprick-based Dengue virus RDT, which will distinguish between the four serotypes.
Building on two long-standing tuberculosis (TB) and HIV patient cohorts in Tanzania and collective expertise by Swiss TPH, the University Hospital Basel (USB) and Ifakara Health Institute (IHI) in pathogen genomics, we have established an innovative integrated pathogen-sequencing platform for HIV-, TB-, antimicrobial resistance (AMR)-related issues, and zoonotic disease pathogen discovery. The platform has strengthened integrated care and diagnostics in Tanzania by providing sequence-guided patient treatment and disease surveillance. It has also created capacity for characterizing emerging pathogens and AMR.

Daniel Paris
Associate Professor, MD, PhD, DTMH, Prof. Dr. med.
Group Leader
+41612848123
daniel.paris@swisstph.ch
Integrated Diagnostics and Sequencing Platform (IDSP) for Emerging and Re-emerging Pathogens and Antimicrobial Resistance in Tanzania
Project overview
In the IDSP project we have built an innovative integrated diagnostics and sequencing platform for the characterization and surveillance of emerging and re-emerging pathogens and antimicrobial resistance development in Tanzania. The project has a strong regional capacity development component with network formation and training of local staff on clinically relevant expertise.
Leveraging excellent pre-existing infrastructure and long-standing productive partnerships, we have maximized synergies by establishing a pathogen-sequencing facility, which serves different clinics and research sites in rural and urban Tanzania. Building up networks of expertise within the country and providing training opportunities for young staff are a central feature of this project. Ultimately, this project aids in improving patient care in Tanzania, enabling the health system to respond more effectively to emerging and re-emerging pathogens and evolving drug resistance, and further promotes innovative science and capacity enhancement in the country.
Goal and research objectives
Building on two long-standing tuberculosis (TB) and HIV patient cohorts in Tanzania and collective expertise by the Swiss TPH, the University Hospital Basel (USB) and Ifakara Health Institute (IHI) in pathogen genomics, we have established an innovative integrated pathogen-sequencing platform for HIV-, tuberculosis (TB)- and antimicrobial resistance (AMR)-related issues, as well as pathogen discovery for emerging and reemerging zoonotic diseases. This platform has:
- strengthened integrated care and diagnostics in Tanzania by providing sequence-guided patient treatment and disease surveillance.
- established capacity for characterizing emerging pathogens and AMR;
- trained diagnostic personnel in molecular diagnostics and pathogen sequencing for AMR determination and improved patient management; and
- enhanced collaboration networks within Tanzania to build representative data collections supporting national health strategies (i.e., National AIDS control Program (NACP), National Tuberculosis and Leprosy Program (NTLP), Ministry of Health and other research groups).
Key problems addressed
- Currently, there are few facilities to detect and characterize emerging pathogens,
- AMR is a highly relevant problem in the management of HIV, TB and malaria patients globally, but currently existing capacity and infrastructure in Tanzania do not allow large-scale detection of AMR for these diseases.
- Detecting transmission hot-spots is crucial for the control and surveillance of infectious diseases, such as TB and HIV, but gene sequencing facilities in Tanzania are scarce.
- There is a significant lack of teaching and training facilities in Tanzania for young medical staff and researchers in the use of molecular tools for routine medical work and clinical research.


Selected Publications
Abongomera C, Keidar O, Paris D.H. Häufig auftretende Krankheiten nach Herkunfts- und Transitländern. In: Exadaktylos A,Keidar O,Srivastava D, eds. Migrations- und Flüchtlingsmedizin, 97-99. Bern: Hogrefe Verlag, 2025
Abongomera C, Labhardt N.D, Paris D.H, Burgener-Gasser A.V. Sexuell übertragbare Infektionen. In: Exadaktylos A,Keidar O,Srivastava D, eds. Migrations- und Flüchtlingsmedizin, 235-244. Bern: Hogrefe Verlag, 2025
Abongomera C, Labhardt N.D, Paris D.H, Burgener-Gasser A.V. Hepatitis B (HBV). In: Exadaktylos A,Keidar O,Srivastava D, eds. Migrations- und Flüchtlingsmedizin, 271-274. Bern: Hogrefe Verlag, 2025
Abongomera C, Labhardt N.D, Paris D.H, Burgener-Gasser A.V. Hepatitis C (HCV). In: Exadaktylos A,Keidar O,Srivastava D, eds. Migrations- und Flüchtlingsmedizin, 274-277. Bern: Hogrefe Verlag, 2025
Abongomera C, Labhardt N.D, Paris D.H, Burgener-Gasser A.V. HIV/AIDS. In: Exadaktylos A,Keidar O,Srivastava D, eds. Migrations- und Flüchtlingsmedizin, 277-284. Bern: Hogrefe Verlag, 2025
Abongomera C, Labhardt N.D, Paris D.H. Strongyloidiasis. In: Exadaktylos A,Keidar O,Srivastava D, eds. Migrations- und Flüchtlingsmedizin, 252-254. Bern: Hogrefe Verlag, 2025
 Charles Abongomera
Charles Abongomera
 Regina Oakley
Regina Oakley